Calling and filtering variants#
In BALSAMIC, various bioinfo tools are integrated for reporting somatic and germline variants summarized in the table below. The choice of these tools differs between the type of analysis; Whole Genome Sequencing (WGS), or Target Genome Analysis (TGA) and Target Genome Analysis (TGA) with UMI-analysis activated.
Variant caller |
Sequencing type |
Analysis type |
Somatic/Germline |
Variant type |
|---|---|---|---|---|
DNAscope |
TGA, WGS |
tumor-normal, tumor-only |
germline |
SNV, InDel |
TNscope |
WGS, TGA (with UMIs activated) |
tumor-normal, tumor-only |
somatic |
SNV, InDel |
VarDict |
TGA |
tumor-normal, tumor-only |
somatic |
SNV, InDel |
Various filters (Pre-call and Post-call filtering) are applied at different levels to report high-confidence variant calls.
Pre-call filtering is where the variant-calling tool decides not to add a variant to the VCF file if the default filters of the variant-caller did not pass the filter criteria. The set of default filters differs between the various variant-calling algorithms.
To know more about the pre-call filters used by the variant callers, please have a look at the VCF header of the particular variant-calling results. For example:
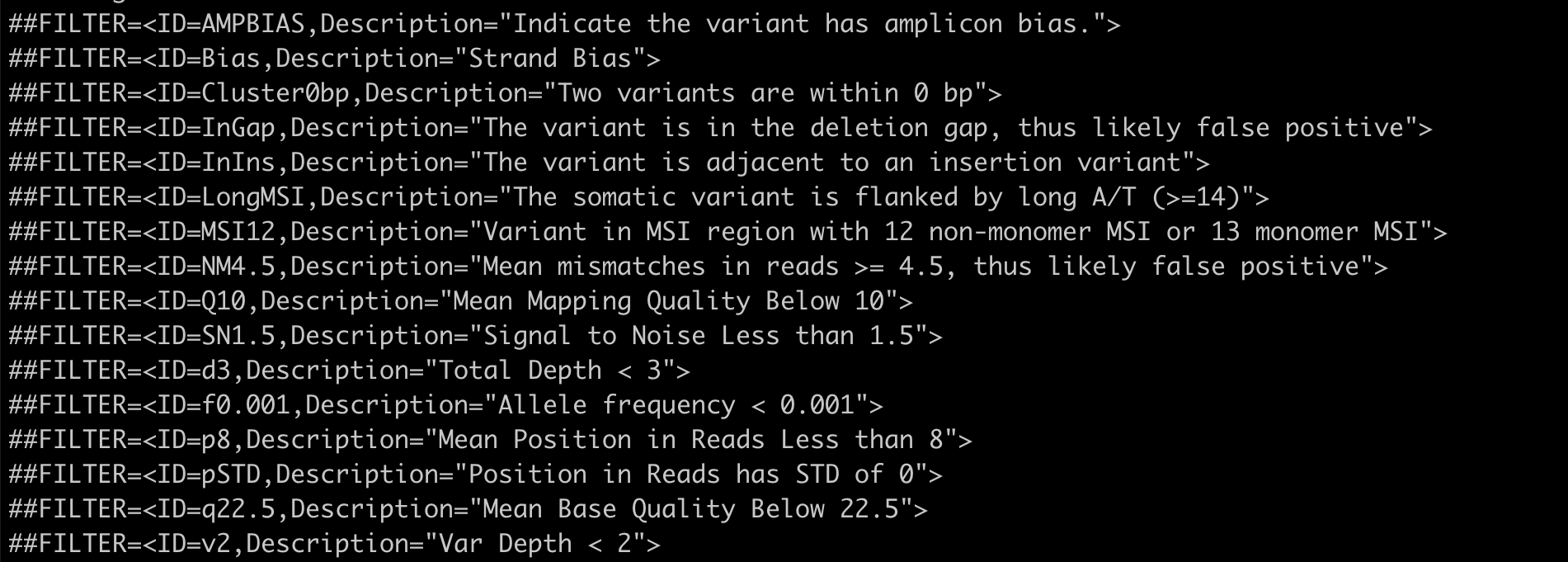
Pre-call filters applied by the Vardict variant-caller is listed in the VCF header.#
In the VCF file, the FILTER status is PASS if this position has passed all filters, i.e., a call is made at this position. Contrary, if the site has not passed any of the filters, a semicolon-separated list of those failed filter(s) will be appended to the FILTER column instead of PASS. E.g., p8;pSTD might indicate that at this site, the mean position in reads is less than 8, and the position in reads has a standard deviation of 0.
Note: In BALSAMIC, this VCF file is named as `*.<research/clinical>.vcf.gz` (eg: `SNV.somatic.<CASE_ID>.vardict.<research/clinical>.vcf.gz`)
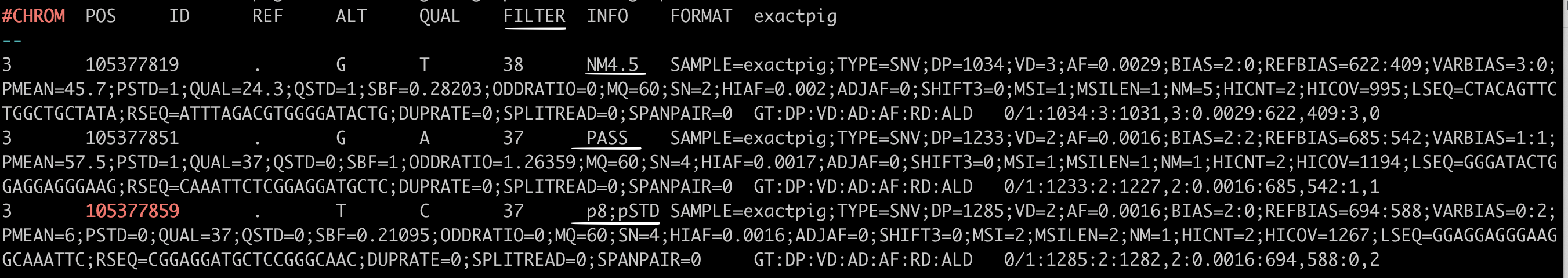
Vardict Variant calls with different ‘FILTER’ status underlined in white line (NM4.5, PASS, p8;pSTD)#
Post-call filtering is where a variant is further filtered with quality, depth, VAF, etc., with more stringent thresholds.
For Post-call filtering, in BALSAMIC we have applied various filtering criteria (Vardict_filtering, TNscope filtering (Tumor_normal) ) depending on the analysis-type (TGS/WGS) and sample-type(tumor-only/tumor-normal).
Note: In BALSAMIC, this VCF file is named as `*.<research/clinical>.filtered.vcf.gz` (eg: `SNV.somatic.<CASE_ID>.vardict.<research/clinical>.filtered.vcf.gz`)
Only those variants that fulfill the pre-call and post-call filters are scored as PASS in the STATUS column of the VCF file. We filter those PASS variants and deliver a final list of variants to the customer either via Scout or Caesar
Note: In BALSAMIC, this VCF file is named as `*.<research/clinical>.filtered.pass.vcf.gz` (eg: `SNV.somatic.<CASE_ID>.vardict.<research/clinical>.filtered.pass.vcf.gz`)
VCF file name |
Description |
Delivered to the customer |
|---|---|---|
.vcf.gz |
Unannotated VCF file with pre-call filters included in the STATUS column |
Yes (Caesar) |
.<research/clinical>.vcf.gz |
Annotated VCF file with pre-call filters included in the STATUS column |
No |
.<research/clinical>.filtered.pass.vcf.gz |
Annotated and filtered VCF file by excluding all filters that did not meet the pre and post-call filter criteria. Includes only variants with the PASS STATUS |
Yes (Caesar and Scout) |
Targeted Genome Analysis#
Somatic Callers for reporting SNVs/INDELS#
Vardict#
Vardict is a sensitive variant caller used for both tumor-only and tumor-normal variant calling. The results of Vardict variant calling are further post-filtered based on several criteria (Vardict_filtering) to retrieve high-confidence variant calls. These high-confidence variant calls are the final list of variants uploaded to Scout or available in the delivered VCF file in Caesar.
There are two slightly different post-processing filters activated depending on if the sample is an exome or a smaller panel as these tend to have very different sequencing depths.
Vardict_filtering#
Following is the set of criteria applied for filtering vardict results. It is used for both tumor-normal and tumor-only samples.
Post-call Quality Filters for panels
Mean Mapping Quality (MQ): Refers to the root mean square (RMS) mapping quality of all the reads spanning the given variant site.
MQ >= 30
Total Depth (DP): Refers to the overall read depth supporting the called variant.
DP >= 100
Variant depth (VD): Total reads supporting the ALT allele
VD >= 5
Allelic Frequency (AF): Fraction of the reads supporting the alternate allele
Minimum AF >= 0.007
Post-call Quality Filters for exomes
Mean Mapping Quality (MQ): Refers to the root mean square (RMS) mapping quality of all the reads spanning the given variant site.
MQ >= 30
Total Depth (DP): Refers to the overall read depth supporting the called variant.
DP >= 20
Variant depth (VD): Total reads supporting the ALT allele
VD >= 5
Allelic Frequency (AF): Fraction of the reads supporting the alternate allele
Minimum AF >= 0.007
Note: Additionally, the variant is excluded for tumor-normal cases if marked as ‘germline’ in the `STATUS` column of the VCF file.
Attention: BALSAMIC <= v8.2.7 uses minimum AF 1% (0.01). From Balsamic v8.2.8, minimum VAF is changed to 0.7% (0.007)
Post-call Observation database Filters
GNOMADAF_POPMAX: Maximum Allele Frequency across populations
GNOMADAF_popmax <= 0.005 (or) GNOMADAF_popmax == "."
SWEGENAF: SweGen Allele Frequency
SWEGENAF <= 0.01 (or) SWEGENAF == "."
Frq: Frequency of observation of the variants from normal Clinical samples
Frq <= 0.01 (or) Frq == "."
Target Genome Analysis with UMI’s into account#
Sentieon’s TNscope#
UMI workflow performs the variant calling of SNVs/INDELS using the TNscope algorithm from UMI consensus-called reads. The following filter applies for both tumor-normal and tumor-only samples.
Pre-call Filters
minreads: Filtering of consensus called reads based on the minimum reads supporting each UMI tag group
minreads = 3,1,1
It means that at least 3 read-pairs need to support the UMI-group (based on the UMI-tag and the aligned genomic positions), and with at least 1 read-pair from each strand (F1R2 and F2R1).
min_init_tumor_lod: Initial Log odds for the that the variant exists in the tumor.
min_init_tumor_lod = 0.5
min_tumor_lod: Minimum log odds in the final call of variant in the tumor.
min_tumor_lod = 4.0
min_tumor_allele_frac: Set the minimum tumor AF to be considered as potential variant site.
min_tumor_allele_frac = 0.0005
interval_padding: Adding an extra 100bp to each end of the target region in the bed file before variant calling.
interval_padding = 100
Post-call Quality Filters
alt_allele_in_normal: Default filter set by TNscope was considered too stringent in filtering tumor in normal and is removed.
bcftools annotate -x FILTER/alt_allele_in_normal
Relative tumor AF in normal: Allows for maximum Tumor-In-Normal-Contamination of 30%.
excludes variant if: AF(normal) / AF(tumor) > 0.3
Post-call Observation database Filters
GNOMADAF_POPMAX: Maximum Allele Frequency across populations
GNOMADAF_popmax <= 0.02 (or) GNOMADAF_popmax == "."
SWEGENAF: SweGen Allele Frequency
SWEGENAF <= 0.01 (or) SWEGENAF == "."
Frq: Frequency of observation of the variants from normal Clinical samples
Frq <= 0.01 (or) Frq == "."
The variants scored as PASS or triallelic_sites are included in the final vcf file (SNV.somatic.<CASE_ID>.tnscope.<research/clinical>.filtered.pass.vcf.gz).
Whole Genome Sequencing (WGS)#
Sentieon’s TNscope#
BALSAMIC utilizes the TNscope algorithm for calling somatic SNVs and INDELS in WGS samples. The TNscope algorithm performs the somatic variant calling on the tumor-normal or the tumor-only samples.
TNscope filtering (Tumor_normal)#
Pre-call Filters
Apply TNscope trained MachineLearning Model: Sets MLrejected on variants with ML_PROB below 0.32.
- ::
ML model: SentieonTNscopeModel_GiAB_HighAF_LowFP-201711.05.model is applied
min_init_tumor_lod: Initial Log odds for the that the variant exists in the tumor.
min_init_tumor_lod = 1
min_tumor_lod: Minimum log odds in the final call of variant in the tumor.
min_tumor_lod = 8
min_init_normal_lod: Initial Log odds for the that the variant exists in the normal.
min_init_normal_lod = 0.5
min_normal_lod: Minimum log odds in the final call of variant in the normal.
min_normal_lod = 1
Post-call Quality Filters
Total Depth (DP): Refers to the overall read depth from all target samples supporting the variant call
DP(tumor) >= 10 (or) DP(normal) >= 10
Allelic Depth (AD): Total reads supporting the ALT allele in the tumor sample
AD(tumor) >= 3
Allelic Frequency (AF): Fraction of the reads supporting the alternate allele
Minimum AF(tumor) >= 0.05
alt_allele_in_normal: Default filter set by TNscope was considered too stringent in filtering tumor in normal and is removed.
bcftools annotate -x FILTER/alt_allele_in_normal
Relative tumor AF in normal: Allows for maximum Tumor-In-Normal-Contamination of 30%.
excludes variant if: AF(normal) / AF(tumor) > 0.3
Post-call Observation database Filters
GNOMADAF_POPMAX: Maximum Allele Frequency across populations
GNOMADAF_popmax <= 0.001 (or) GNOMADAF_popmax == "."
SWEGENAF <= 0.01 (or) SWEGENAF == "."
Frq: Frequency of observation of the variants from normal Clinical samples
Frq <= 0.01 (or) Frq == "."
The variants scored as PASS or triallelic_sites are included in the final vcf file (SNV.somatic.<CASE_ID>.tnscope.<research/clinical>.filtered.pass.vcf.gz).
TNscope filtering (tumor_only)#
Pre-call Filters
min_init_tumor_lod: Initial Log odds for the that the variant exists in the tumor.
min_init_tumor_lod = 1
min_tumor_lod: Minimum log odds in the final call of variant in the tumor.
min_tumor_lod = 8
The somatic variants in TNscope raw VCF file (SNV.somatic.<CASE_ID>.tnscope.all.vcf.gz) are filtered out for the genomic regions that are not reliable (eg: centromeric regions, non-chromosome contigs) to enhance the computation time. This WGS interval region file is collected from gatk_bundles gs://gatk-legacy-bundles/b37/wgs_calling_regions.v1.interval_list.
Post-call Quality Filters
Total Depth (DP): Refers to the overall read depth supporting the variant call
DP(tumor) >= 10
Allelic Depth (AD): Total reads supporting the ALT allele in the tumor sample
AD(tumor) > 3
Allelic Frequency (AF): Fraction of the reads supporting the alternate allele
Minimum AF(tumor) > 0.05
SUM(QSS)/SUM(AD) >= 20
Read Counts: Count of reads in a given (F1R2, F2R1) pair orientation supporting the alternate allele and reference alleles
ALT_F1R2 > 0, ALT_F2R1 > 0
REF_F1R2 > 0, REF_F2R1 > 0
SOR: Symmetric Odds Ratio of 2x2 contingency table to detect strand bias
SOR < 3
Post-call Observation database Filters
GNOMADAF_POPMAX: Maximum Allele Frequency across populations
GNOMADAF_popmax <= 0.001 (or) GNOMADAF_popmax == "."
Normalized base quality scores: The sum of base quality scores for each allele (QSS) is divided by the allelic depth of alt and ref alleles (AD)
SWEGENAF <= 0.01 (or) SWEGENAF == "."
Frq: Frequency of observation of the variants from normal Clinical samples
Frq <= 0.01 (or) Frq == "."
The variants scored as PASS or triallelic_sites are included in the final vcf file (SNV.somatic.<CASE_ID>.tnscope.<research/clinical>.filtered.pass.vcf.gz).
Attention: BALSAMIC <= v8.2.10 uses GNOMAD_popmax <= 0.005. From Balsamic v9.0.0, this settings is changed to 0.02, to reduce the stringency. BALSAMIC >= v11.0.0 removes unmapped reads from the bam and cram files for all the workflows. BALSAMIC >= v13.0.0 keeps unmapped reads in bam and cram files for all the workflows.